1. Introduction
In this tutorial, we’ll focus on bias estimation and confidence intervals via bootstrapping in one-sample settings. We’ll explain how and why bootstrap works and show how to implement the percentile and reversed bootstrapped confidence intervals in Python.
2. When Is Bootstrap Useful?
Let’s say we want to estimate the efficiency of an algorithm. So, we define 50 various inputs, execute our algorithm, and record the execution times. To make the average estimate more robust to outliers (which represent too easy and too hard inputs), we drop the bottom 10% and top 10% values and compute the mean of the inner 80%. This is an example of a trimmed mean.
To find the confidence interval around the computed value or to make other statistical inferences, we need to know the distribution of the trimmed mean as a statistic (a random variable). By the central limit theorem, the mean of a sample is asymptotically normally distributed, but that’s not necessarily the case with the mean of a trimmed sample.
The bootstrap technique is useful when we don’t know the distribution of a sample statistic because analytical derivation is too complex or impossible. Bootstrap allows us to approximate the distribution and use it for statistical inference.
3. Bootstrap
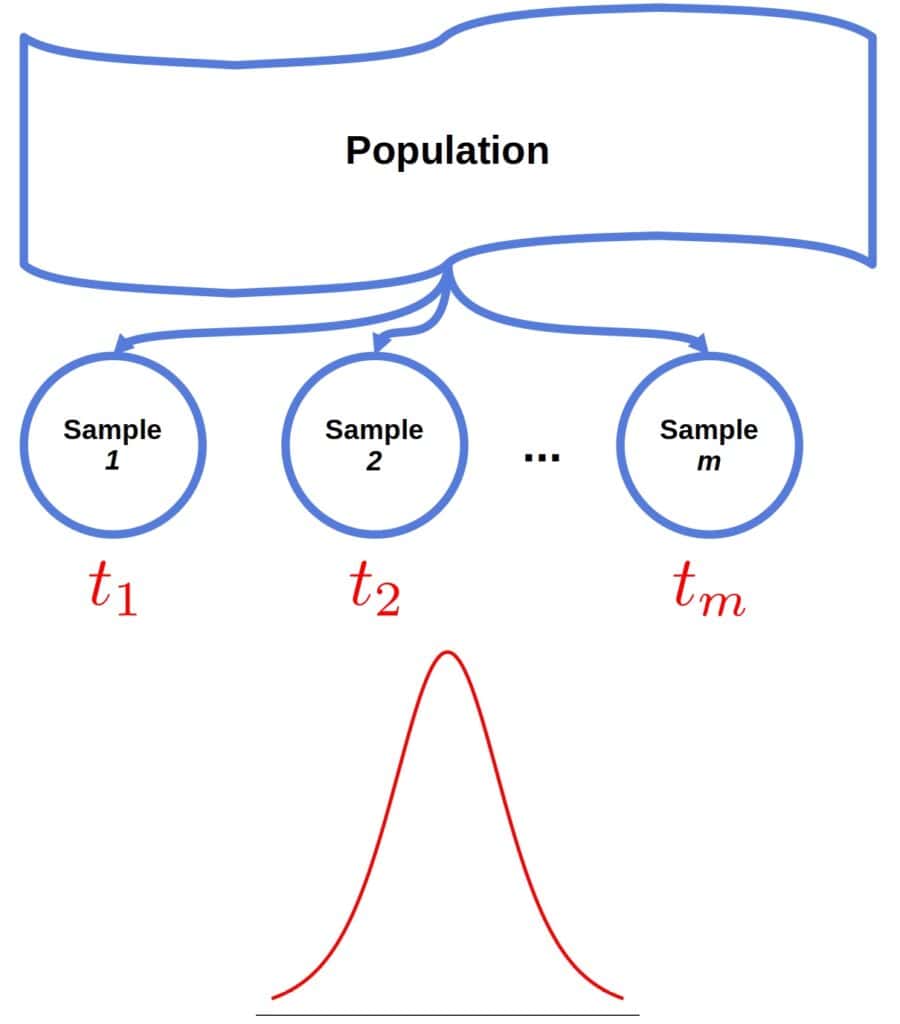
If we could draw sufficiently many samples from the underlying population and calculate a statistic for each sample, we could approximate the statistic’s sampling distribution:
Let’s say we repeated the run-time experiment 1000 times. In replication, we ran the algorithm 50 times and computed the trimmed mean. That way, we would get an empirical approximation of the trimmed mean’s distribution.
However, resampling from the population may be hard or impossible. For instance, the algorithm may be expensive to execute because it requires extensive computational resources.
3.1. The Bootstrap Idea
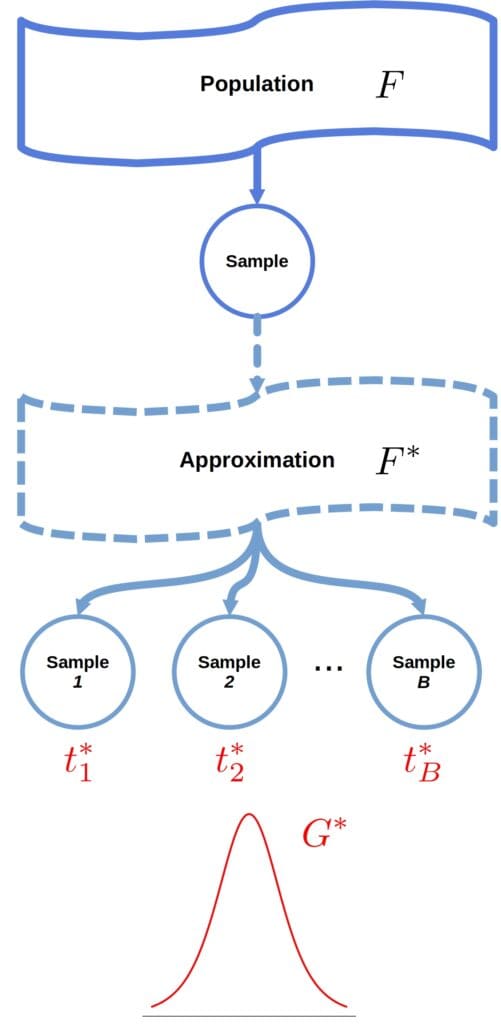
In bootstrapping, we define an approximate mathematical model (i.e., distribution) from which we can sample straightforwardly. So, instead of resampling from the true population (a distribution with CDF ), we draw a large number
of resamples (called bootstrap samples) from an approximation
:
This way, we compute , the CDF of the statistic
, where
. Under certain conditions, one of which is that the approximation
is good enough, we can expect
to have the same shape and spread as
, the CDF of
when
(we would get
if we could resample from
at will).
Note that if we can derive the distribution analytically, we don’t need
bootstrap samples. However, that’s not the case for most statistics. In practice, we set
to a high enough value and approximate
by sampling.
3.2. Types of Bootstrap
We can categorize bootstrap techniques depending on how they obtain .
Parametric bootstrap assumes the family of
, estimates its parameters from the initial sample
, and defines
as the CDF in family
with those parameters. For example, we can assume that
is the CDF of an exponential distribution (with the density
), and use
to find an estimate
of
. Then,
will have the density
.
Smoothed bootstrap uses kernel density estimation to arrive at .
Nonparametric bootstrap assumes nothing about the shape or family of and uses the empirical CDF based on
as \boldsymbol{F^*}. Here, we draw from
by sampling from
with replacement.
4. Bootstrap and Statistical Inference
4.1. Bootstrap Approximates the Error Distribution
Although we expect their shapes and spread to be similar, the means of and
need not be identical. The bootstrapped
values will fluctuate around the mean of
. Let’s denote it as
, where
is the potential bias, and
is the population value of interest with respect to
. This mean isn’t necessarily equal to the mean of
(
), where
is the value of
in the actual population.
Therefore, another condition for the similarity of shapes (and spread) is that they don’t depend on the locations, i.e., that the moments of don’t depend on the mean of
and that the moments of
don’t depend on the mean of
.
However, all said and done, even if the shapes are similar, the locations can and usually differ.
Then, why is useful? The approximation
should be such that we can easily compute
. Subtracting
from a
-distributed random variable, we get a model for the error of
:
Because the shapes are the same (or similar enough), the error of follows a similar distribution:
We can use the distribution of to analyze the bias of
and construct the confidence intervals for
.
Let be the original sample (from
) and let
be the original estimate of
.
In non-parametric bootstrap, is the ECDF
based on the original sample
, so
.
4.2. Bias
The value estimates the bias with respect to
.
Under the assumption that if , we’ll have
, so we can use
as the bias estimate for
in the original population.
If it’s non-zero, we can consider a biased statistic. Otherwise, we say it’s unbiased or a consistent estimator.
We may be tempted to use the bias estimate to correct , but such corrections may do more harm than good because bias estimates can be unstable.
4.3. Reversed Confidence Intervals
Let’s say the statistic is unbiased. Let the and
quantiles of
be
and
. In practice, we sort the
values and use those at positions
and
. Then:
From there, we get:
So, the interval for is:
Since the quantiles are reversed in the final interval, we call these bootstrap intervals reversed.
4.4. Percentile Confidence Intervals
There are other bootstrap intervals. We’ll cover the construction of percentile confidence intervals.
The percentile method has an even stronger assumption that in shape, spread, and location. So, in this method, we treat the quantiles
and
of
as the corresponding quantiles of
.
Therefore, the percentile interval (for ) is:
Reversed and percentile intervals are easy to compute but fail if their assumptions aren’t met.
4.5. Bootstrap Assumptions
Bootstrap assumes that the bootstrapped distribution is similar enough to the actual distribution
of the statistic of interest.
For that reasoning to hold, must approximate
well, and
should be robust to small changes in its argument.
The former condition is there because we can’t use as a proxy for
if it isn’t precise. The latter condition ensures that small deviations of
from
don’t translate to large deviations of
from
(this property is known as smoothness). If these conditions aren’t met, bootstrap won’t be useful.
There may be additional assumptions depending on the bootstrap technique. For example, the confidence-interval methods we discussed assume that the shape and spread of and
don’t depend on their location parameters.
5. Python Implementation
Let’s implement reversed and percentile interval methods in Python.
We’ll use nonparametric bootstrap, which means we’ll take bootstrap samples from the original sample with replacement.
We’ll need numpy and scipy:
import numpy
import scipy.stats
5.1. Code for Reversed Intervals
Here’s the code for reversed intervals:
def bootstrap_reversed_ci(x, alpha, B):
# Compute the sample statistic
t = scipy.stats.trim_mean(x, 0.1)
# Draw bootstrap samples
n = len(x)
x_bootstrap = np.random.choice(x, size=(B, n), replace=True)
# Compute the bootstrap statistics
t_bootstrap = scipy.stats.trim_mean(x_bootstrap, 0.1, axis=1)
# Find the error distribution
# (In non-parametric bootstrap, the population mean of t_bootstrap is t.)
err_bootstrap = t_bootstrap - t
# Find the quantiles
q_lower, q_upper = np.quantile(err_bootstrap, (alpha/2, 1 - alpha/2))
return t, (t - q_upper, t - q_lower)
Here, x_bootstrap is a matrix. By specifying axis=1 in scipy.stats.trim_mean(), we calculate the trimmed mean for each row and obtain a
numpy array. This and other calculations are vectorized to improve performance.
The array err_bootstrap holds the differences for
. Since we use the nonparametric bootstrap,
is the trimmed mean of the original sample x.
Finally, we use the alpha/2 and (1-alpha/2) quantiles of err_bootstrap to define the confidence interval and return it alongside the sample trimmed mean.
5.2. Code for Percentile Intervals
Here’s the code for percentile intervals:
def bootstrap_percentile_ci(x, alpha, B):
# Compute the sample statistic
t = scipy.stats.trim_mean(x, 0.1)
# Draw bootstrap samples
n = len(x)
x_bootstrap = np.random.choice(x, size=(B, n), replace=True)
# Compute the bootstrap statistics
t_bootstrap = scipy.stats.trim_mean(x_bootstrap, 0.1, axis=1)
# Find the quantiles
q_lower, q_upper = np.quantile(t_bootstrap, (alpha/2, 1 - alpha/2))
return t, (q_lower, q_upper)
Here, we also perform vectorized calculations. The main difference is that we don’t compute the error distribution. Instead, we use the alpha/2 and (1 – alpha/2) quantiles of the array t_bootstrap of the bootstrapped statistics as the lower and upper boundaries of the confidence interval.
5.3. Results
Let’s see how to use these functions. As an example, we draw the original sample from an exponential distribution , but any other distribution is suitable. We draw
bootstrap samples and aim for the coverage of
:
x = scipy.stats.expon.rvs(scale=2, size=50)
print(bootstrap_reversed_ci(x, 0.05, 1000))
print(bootstrap_percentile_ci(x, 0.05, 1000))
Here’s an example output:
1.685 (1.237, 2.0355)
1.685 (1.2081, 2.0595)
We see the sample trimmed mean and the reversed and percentile intervals. Both methods produced similar results and captured the true trimmed mean of approximately 1.6615.
To use these bootstrap functions with another statistic, we need to replace scipy.stats.trim_mean.
6. Conclusion
In this article, we explained the ideas behind bootstrap in statistics and showed how to estimate the bias and compute confidence intervals.
Bootstrap is useful when we don’t know the sampling distribution of a statistic, but it can fail if its assumptions aren’t met.